Dealing with factors
By Deependra Dhakal in R factor blog
May 18, 2017
A factor is an headache
I have a dataset, cleaning which has been a pain lately. I’m going to use 20 observations of the imported dataset in this post to demonstrate how pathetically have I been advancing with it.
| plot | jan_23_2017 | jan_26_2017 | jan_29_2017 | feb_02_2017 |
|---|---|---|---|---|
| 1 | 0 | 0 | b | am |
| 2 | f | s | 1p | 10p |
| 3 | b | a | sm | 3p |
| 4 | b | b | ap | 2 |
| 5 | 0 | b | bp | s |
| 6 | b | a | sp | 3 |
Providing it a context, the columns represent multiple observations of same variable at different dates, as apparent from the column names.
One can observe from the str(head_boot) that the dataframe has only 20 observations. Had a larger dataframe been subsetted, the factors with their usual nuisance carry over the all the levels, even currently unused ones. To demonstrate this, below I present how many factor levels the original dataframe had and what changes after a subset (slice, filter have same feature) operation.
# original dataframe
head_boot %>%
select_if(is.factor) %>%
map_int(function(x)length(levels(x)))
## jan_23_2017 jan_26_2017 jan_29_2017 feb_02_2017
## 5 6 14 15
# subsetted to obtain a dataframe where one factors has either "s" or "0" levels only
subset.data.frame(head_boot,
subset = head_boot$jan_23_2017 %in% c("s", "0"),
select = sapply(head_boot, is.factor)) %>%
map_int(function(x)length(levels(x)))
## jan_23_2017 jan_26_2017 jan_29_2017 feb_02_2017
## 5 6 14 15
All factor levels are retained !
A prophylactic measure is avoiding at all importing of character data columns as a factor. This could be done using setup option: options(stringsAsFactors = FALSE) ahead of import. Still, after the import many options exist. Some of the best alternatives out are listed below:
-
Use
subsetted_df$factor_col[, drop=TRUE], where a vector of factor values is indexed with drop argument. -
Convert factor columns of subsetted dataframe first to character and then back to factors class.
-
Use
forcats::fct_drop(f)for more reliable NA handling -
Use
droplevels()ordroplevels.data.frame()
To give a realistic feel to data, factor levels may be recoded or relabelled with more informative labels.
suit_labels <- factor(c("before_boot_n_boot", "heading", "anthesis"),
levels = c("before_boot_n_boot", "heading", "anthesis"),
ordered = T)
# what are all the levels
head_boot_lvl <- head_boot %>%
select_if(is.factor) %>%
map(levels) %>%
unlist() %>%
unique()
# create all combinations of factor levels of the dataset
# purrr::cross_df preserves the order, tidyr::crossing doesn't
possible_hb_lvl <- cross_df(list(post = factor(c("m", "", "p"), ordered = TRUE),
pre = factor(c("b", "f", "a", "s", "1", "2",
"3", "4", "10", "11", "12"), ordered = TRUE))) %>%
select(rev(everything())) %>%
# arrange(match(post, c("m", "", "p"))) %>% # don't need this ordering
unite(col = "pre_post", sep = "") %>%
add_row(pre_post = "0", .before = 1) %>%
mutate(pre_post = factor(pre_post, levels = pre_post, ordered = TRUE))
# mutate original dataframe to represent ordinal factors
head_boot <- head_boot %>%
mutate_if(is.factor, function(x)factor(x, levels = possible_hb_lvl$pre_post, labels = possible_hb_lvl$pre_post, ordered = TRUE))
# function to recode factor levels (better to use `fct_recode()`)
fct_minimize <- function(x)case_when(x < "1m" ~ suit_labels[1],
x >= "1m" & x < "10m" ~ suit_labels[2],
x >= "10m" ~ suit_labels[3],
TRUE ~ NA)
# mutate original dataframe with function to recode factor levels
head_boot <- head_boot %>%
mutate_if(is.factor, fct_minimize)
str(head_boot)
## tibble [20 × 5] (S3: tbl_df/tbl/data.frame)
## $ plot : num [1:20] 1 2 3 4 5 6 7 8 9 10 ...
## $ jan_23_2017: Ord.factor w/ 3 levels "before_boot_n_boot"<..: 1 1 1 1 1 1 1 1 1 1 ...
## $ jan_26_2017: Ord.factor w/ 3 levels "before_boot_n_boot"<..: 1 1 1 1 1 1 1 2 1 1 ...
## $ jan_29_2017: Ord.factor w/ 3 levels "before_boot_n_boot"<..: 1 2 1 1 1 1 2 2 2 1 ...
## $ feb_02_2017: Ord.factor w/ 3 levels "before_boot_n_boot"<..: 1 3 2 2 1 2 2 3 2 1 ...
A more telling dataframe looks like the one below.
knitr::kable(head(head_boot), format = "html", align = "c") %>%
kableExtra::kable_styling(bootstrap_options = c("striped", "hover"),
font_size = 10, position = "center") %>%
kableExtra::row_spec(0, bold = TRUE) %>%
kableExtra::column_spec(1, bold = TRUE)
| plot | jan_23_2017 | jan_26_2017 | jan_29_2017 | feb_02_2017 |
|---|---|---|---|---|
| 1 | before_boot_n_boot | before_boot_n_boot | before_boot_n_boot | before_boot_n_boot |
| 2 | before_boot_n_boot | before_boot_n_boot | heading | anthesis |
| 3 | before_boot_n_boot | before_boot_n_boot | before_boot_n_boot | heading |
| 4 | before_boot_n_boot | before_boot_n_boot | before_boot_n_boot | heading |
| 5 | before_boot_n_boot | before_boot_n_boot | before_boot_n_boot | before_boot_n_boot |
| 6 | before_boot_n_boot | before_boot_n_boot | before_boot_n_boot | heading |
Visualizing data
Bar plotting of the ordinal data might reveal interesting insights, so let’s prepare ggplot graphs.
walk2(.x = head_boot %>% select(jan_23_2017:feb_02_2017) %>% colnames(),
.y = as.character(strptime(str_subset(colnames(head_boot), "^\\w{3}_\\d{2}"),
format = "%b_%d_%Y")),
.f = ~print(ggplot(aes(x = get(.x)), data = head_boot) +
geom_bar(position = "dodge", stat = "count", width = .5) +
xlab(.y) +
theme(axis.text.x = element_text(angle = 40, size = 9, vjust = 0.6))))
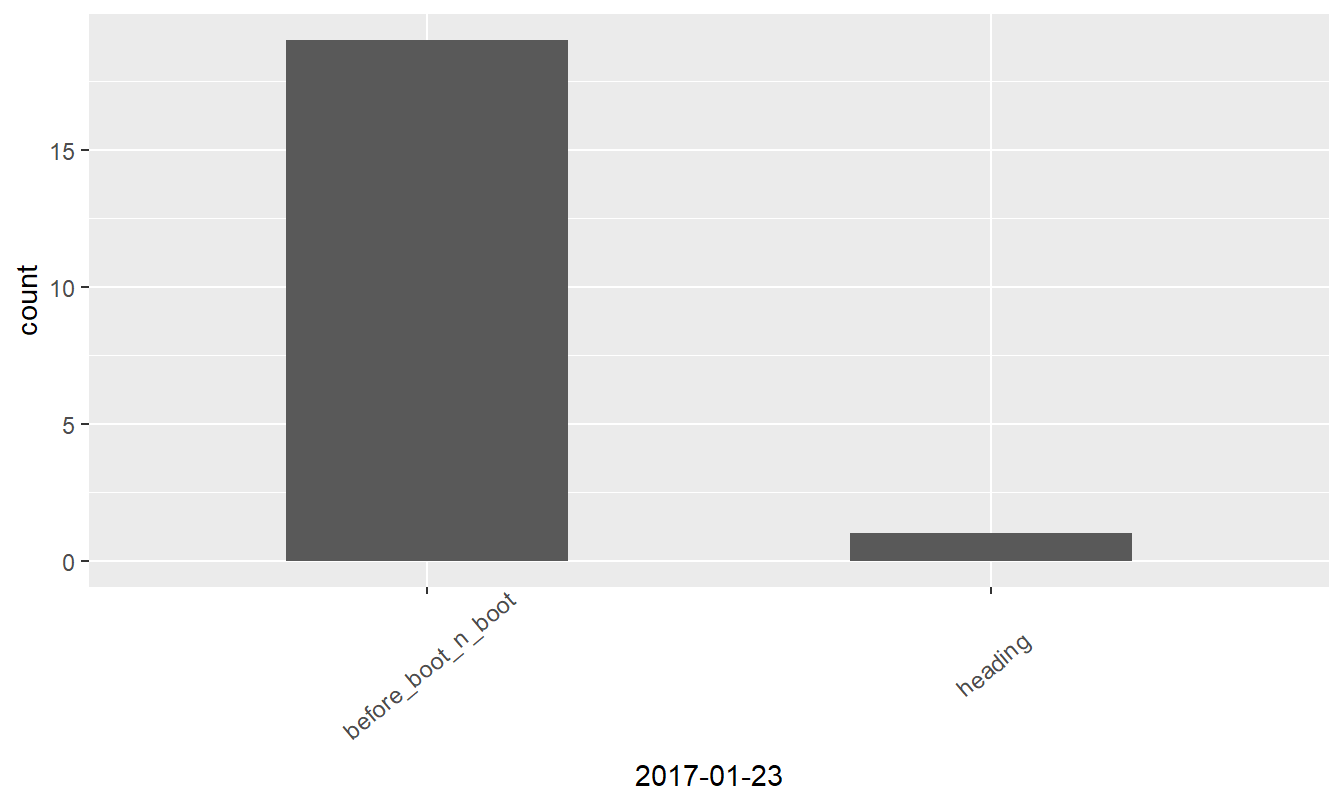
Figure 1: Growth stages at different dates
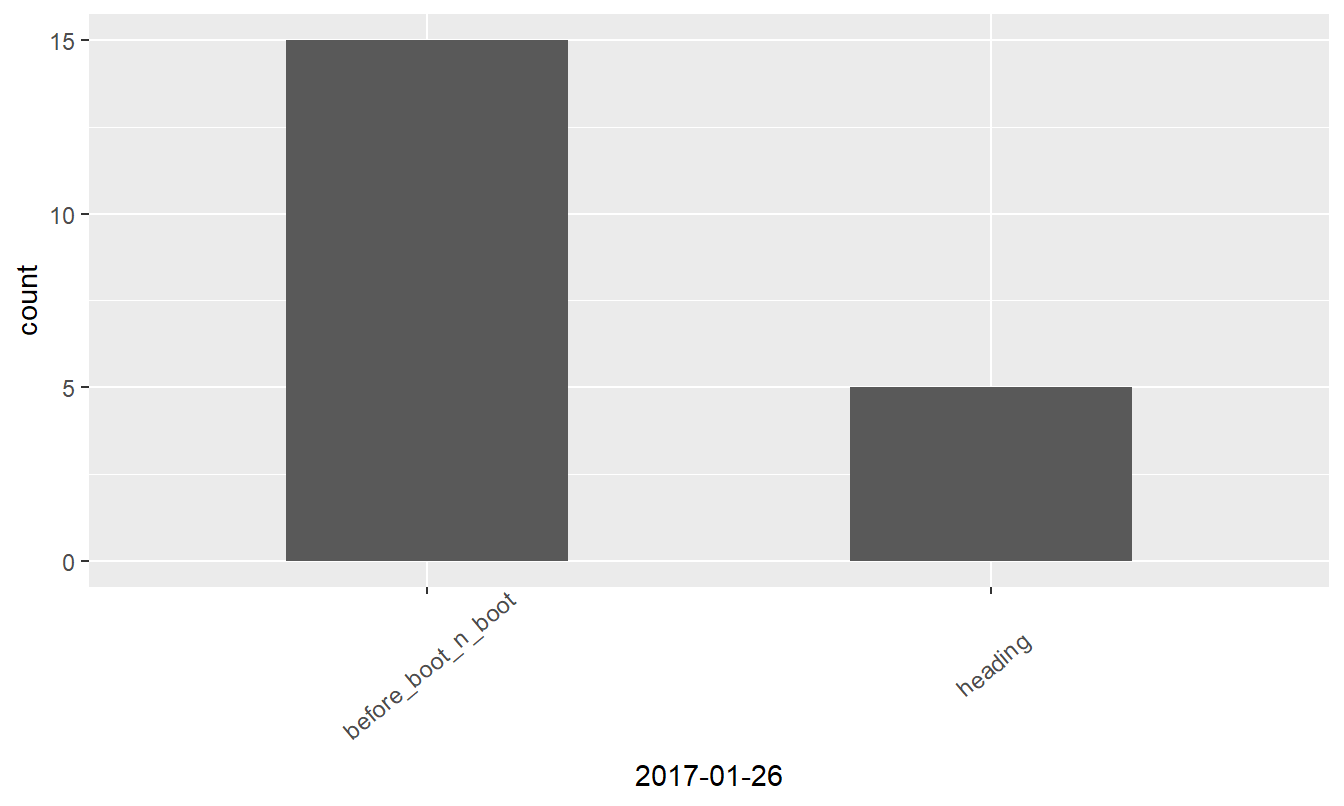
Figure 2: Growth stages at different dates
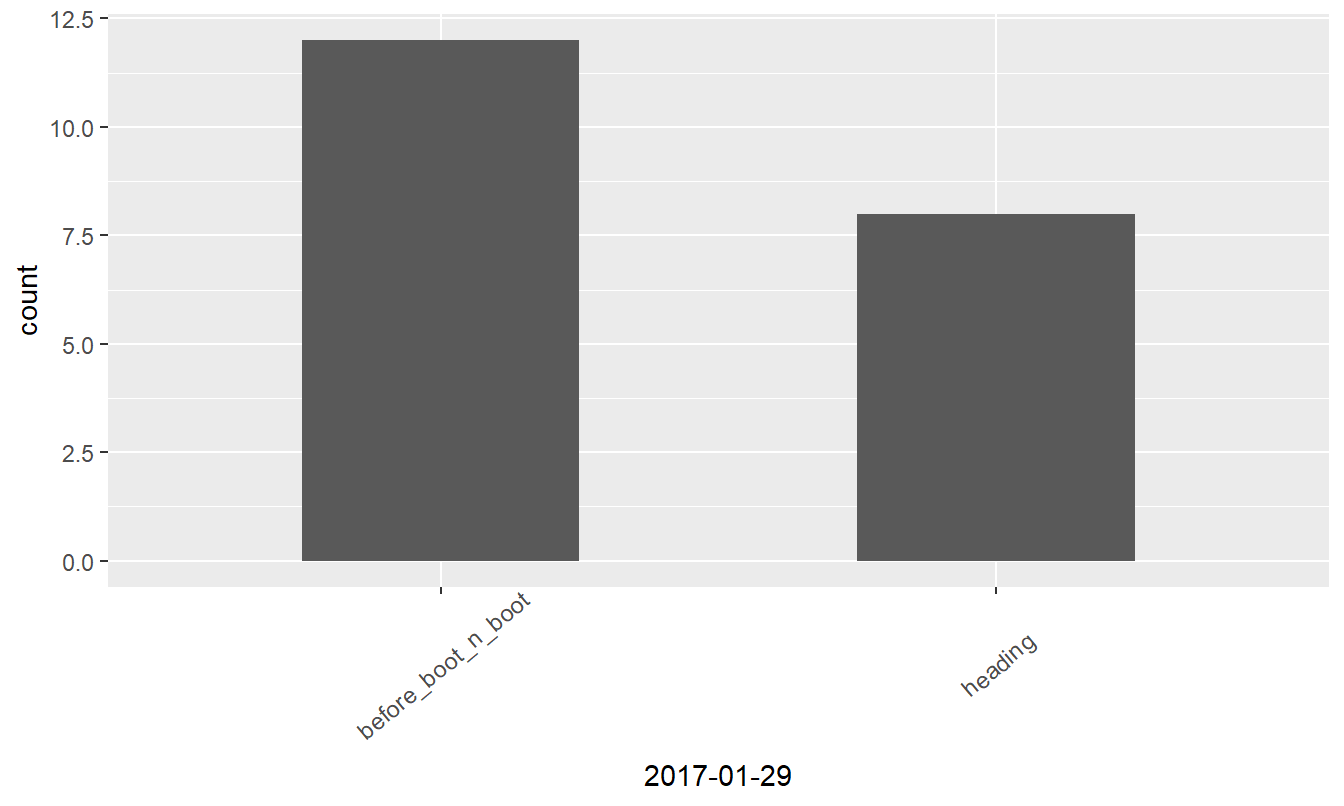
Figure 3: Growth stages at different dates
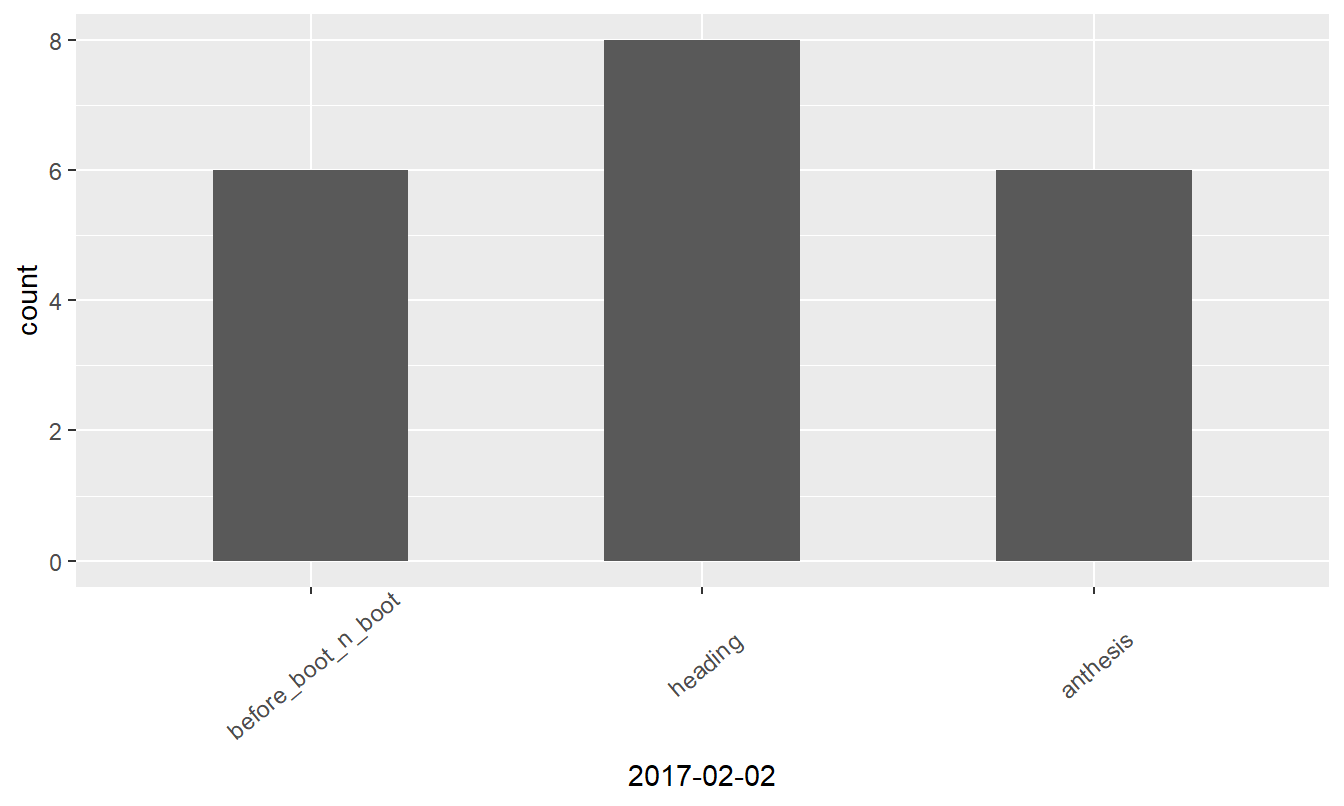
Figure 4: Growth stages at different dates
With dplyr, It’s seemless to obtain a tabular summary of what has been visualized above.
# credit: https://stackoverflow.com/a/46340237/6725057
head_boot %>%
select_if(is.factor) %>%
tidyr::gather(date, stage) %>%
dplyr::group_by(date, stage) %>%
dplyr::count() %>%
dplyr::ungroup() %>%
tidyr::spread(date, n) %>%
knitr::kable(format = "html", align = "c") %>%
kableExtra::kable_styling(bootstrap_options = c("striped", "hover"),
font_size = 10, position = "center") %>%
kableExtra::row_spec(0, bold = TRUE) %>%
kableExtra::column_spec(1, bold = TRUE)
| stage | feb_02_2017 | jan_23_2017 | jan_26_2017 | jan_29_2017 |
|---|---|---|---|---|
| anthesis | 6 | |||
| before_boot_n_boot | 6 | 19 | 15 | 12 |
| heading | 8 | 1 | 5 | 8 |
- Posted on:
- May 18, 2017
- Length:
- 4 minute read, 847 words